|

Project Director
Nigel Martin
Academic Staff
Alex Poulovassilis
S. Hubbard
S. Oliver
S. Embury
N. Paton
C. Goble
R. Stevens
D. Jones
C. Orengo
Research Staff
L. Zamboulis
K. Belhajjame
J. Siepen M. Pentony
R. Apweiler
H. Hermjakob
W. Zhu
C. Taylor
P. Jones
N. Vinod
Project Details
3 years. Funder: BBSRC
Keywords
Bioinformatics
Data Integration
Grid Computing
|
iSPIDER – A Pilot Grid for Integrative Proteomics
ISPIDER is developing an integrated platform of proteome-related resources, using existing standards from proteomics, bioinformatics and e-Science. The project is Grid-enabling existing proteomics data resources, creating new resources, producing middleware technologies for the integration of these resources – including tools for data integration, workflows and data analysis – and producing visualisation and other types of clients for biologist end users.
Experimental proteomics is an essential component for the elucidation of protein biological functions. It involves the study of a set of proteins produced by an organism with the aim of understanding their behaviour under a variety of experimental conditions and environments.
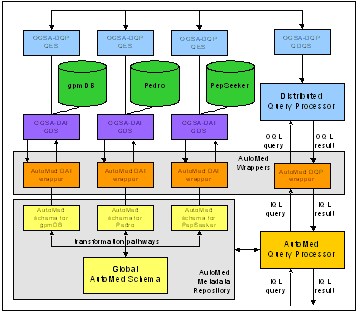
Our approach is based on the interoperation of Grid data access (OGSA-DAI), Grid distributed querying (OGSA-DQP) and data integration (AutoMed) software tools.
OGSA-DAI (http://ww.ogsadai.org.uk/) is an open-source, extendable middleware product exposing data resources on Grids via web services. Efficient querying of OGSA-DAI Grid resources via parallelism is supported by OGSA-DQP (http://ww.ogsadai.org.uk/about/ogsa-dqp), a service-based distributed query processor.
The AutoMed (http://ww.doc.ic.ac.uk) heterogeneous data integration system assists in the transformation and integration of data from different data sources expressed in possibly different data models. This is achieved by defining transformation pathways between schemas.
Transformation pathways between individual proteomics resources, such as gpm DB, and a global schema are defined and stored in the AutoMed Metadata Repository. A query posed on the global schema is submitted to the AutoMed Query Processor, which reformulates it using the transformation pathways into a suitable query for evaluation by the data sources. The query is then optimised and wrapper software translates it from IQL to OQL, the query languages of AutoMed and DQP respectively. DQP evaluates the query by interacting with the data sources via OGSA-DAI services. Results are then combined and transformed in the reverse direction.
http://www.dcs.bbk.ac.uk/~lucas/projects/ispider
|